biostats.pca_plot#
- biostats.pca_plot(data, x, color=None)[source]#
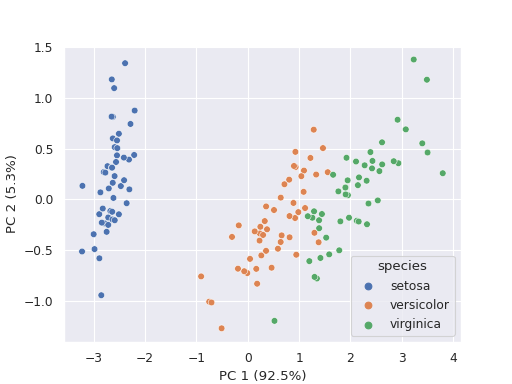
Perform a principle component analysis and draw a scatter plot to show the transformed data.
- Parameters:
- data
pandas.DataFrame The input data. Must contain at least two numeric columns.
- x
list The list of numeric variables to be analyzed.
- color
str The categorical variable specifying groups to be plotted with different colors. Maximum 20 groups. Optional.
- data
- Returns:
- fig
matplotlib.figure.Figure The generated plot.
- fig
See also
fa_plotPerform a factor analysis and draw a scatter plot to show the transformed data.
lda_plotPerform a linear discriminant analysis and draw a scatter plot to show the transformed data.
principal_component_analysisFind the linear combination of a set of variables to manifest the variation of data.
Examples
>>> import biostats as bs >>> import matplotlib.pyplot as plt >>> data = bs.dataset("iris.csv") >>> data sepal_length sepal_width petal_length petal_width species 0 5.1 3.5 1.4 0.2 setosa 1 4.9 3.0 1.4 0.2 setosa 2 4.7 3.2 1.3 0.2 setosa 3 4.6 3.1 1.5 0.2 setosa 4 5.0 3.6 1.4 0.2 setosa .. ... ... ... ... ... 145 6.7 3.0 5.2 2.3 virginica 146 6.3 2.5 5.0 1.9 virginica 147 6.5 3.0 5.2 2.0 virginica 148 6.2 3.4 5.4 2.3 virginica 149 5.9 3.0 5.1 1.8 virginica
We want to perform a principal component analysis and visualize the transformed data.
>>> fig = bs.pca_plot(data=data, x=["sepal_length", "sepal_width", "petal_length" ,"petal_width"], color="species") >>> plt.show()